Labelbox•September 25, 2020
How MIT researchers are using neural nets to automate manual tasks for novel serotonin research
Jungsoo Kim is a PhD graduate student at MIT’s Flavell Lab which focuses on cutting-edge neuroscience research. His team is currently exploring how computation and behavior emerge from neural circuits. Neurons, just like electronics, need to be properly wired in order to function well. Their project investigates how serotonin modulates a large number of neurons, altering the circuit dynamics in the brain. Serotonin supports a wide range of computation and behavior ranging from mood regulation to learning. Bringing new insights at the implementational-level could chart new paths for many research areas such as mental health.
Let’s take a look at how Jungsoo applied deep neural nets as a tool to automate and speed up data processing and analysis for their project.
The role of neural nets and high-quality training data
Training a deep neural network traditionally requires a lot of data. While there has been a lot of discussion around the use of transfer learning to significantly reduce the data need, Jungsoo found that transfer learning was limited for his specific use case because pre-trained networks were trained on the images that had different statistical properties from the specialized datasets they collected. Beyond their training needs, Jungsoo’s team needed high-quality labeled data to benchmark their models.
The key process innovation in their research was that their team was able to successfully automate and replace many of their manual processing pipelines with neural networks (e.g. tracking, segmentation, event/object detection, etc.). In the example we’ll cover below, Jungsoo worked on a model which used object detection and found the labeling process to be a monotonous task that could be vastly sped up with the right tools.
In order to dissect a neural circuit, Jungsoo’s team simultaneously recorded and controlled a large population of neurons. To achieve this, they employed a variety of molecular, optical, and computational tools to image and extract information. As one of those tools, Jungsoo needed a web-based tool so that he could distribute the image annotation task across multiple annotators in the group. After a quick online search and testing out multiple options, he found Labelbox to be the best option with the most intuitive UI, data structures, and API.
Data labeling workflow
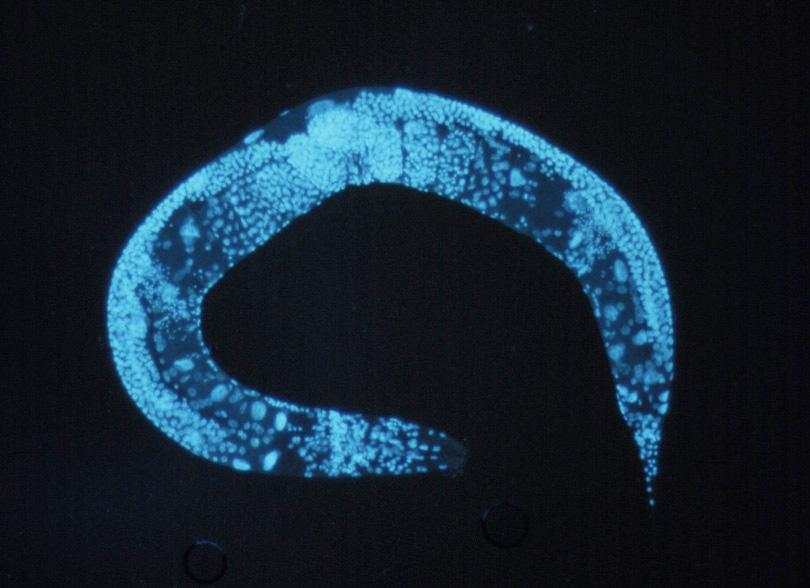
After loading up and signing up for an account Labelbox, Jungsoo set up his project to handle object detection methods so that he could quickly locate the object of freely behaving worms (C. elegans) with the goal of recording and controlling their neural activity. The purpose of the labeling exercise was to identify whether the worm was in the image, and then to mark its centroid. As the images below show, identifying the centroid means having an annotator find the head section of the worm, which typically shows up as a small dot with a slightly dark contrast, located inside the bulb-shaped region (posterior bulb). The classification needed was to identify 1 instance of the object per image (or none), and they labeled 2,000-3,000 images for their initial project.
In the lab, Jungsoo and his team collected terabytes of images and videos every week from behavior cameras and microscopes of these organisms. Historically, they processed the images either manually or with “traditional” computer vision tools (i.e. non-learning based), which he found to be labor-intensive and not robust with varying experimental conditions.
To address any quality concerns labeling errors, Jungsoo set up review workflows in his project so that a third of the images would be labeled at least 3 times. Once the labeling was complete, Jungsoo wrote a script to check the multipass labels against a predetermined criterion and the ground-truth data.
Jungsoo believed that automating many of their processing pipelines with neural nets would speed up their data collection and processing which turned out to yield dividends. Collectively, these models saved their team thousands of hours, which in turn could then be redirected to research and higher-level scientific tasks, as opposed to data preparation and annotation.
Benefits of using Labelbox + Workforce
Jungsoo wanted to share his Labelbox experience so that other researchers and data science teams may benefit from having access to some of the world’s best labelers right within their own Labelbox project.
“Working with Labelbox and the Workforce team was very straightforward. The turnaround was quicker than I expected, typically within overnight or 24 hrs. It was also easy to add more datasets to be labeled to the project. In terms of the labeling quality, I was excited to see that 98% of our multiplass labeled images met predefined quality criterion and the annotation quality was better than I expected.
Another reason behind using Labelbox was having real people that I can contact for feedback and questions. The Workforce team was super helpful and responsive, and it was great to have the project managers. When I had any questions about the labeling process and the product's user interface, I was always able to get the right answers quickly.”
We wanted to thank Jungsoo for sharing his experience and we’re inspired by the work that his lab is doing to advance our scientific understanding of serotonin and mental health.

 All blog posts
All blog posts

